| PICALM |
|---|
 |
| Identifiers |
|---|
| Aliases | PICALM, CALM, CLTH, LAP, phosphatidylinositol binding clathrin assembly protein |
|---|
| External IDs | OMIM: 603025; MGI: 2385902; HomoloGene: 111783; GeneCards: PICALM; OMA:PICALM - orthologs |
|---|
| Gene location (Human) |
|---|
 | | Chr. | Chromosome 11 (human)[1] |
|---|
| | Band | 11q14.2 | Start | 85,957,175 bp[1] |
|---|
| End | 86,069,882 bp[1] |
|---|
|
| Gene location (Mouse) |
|---|
 | | Chr. | Chromosome 7 (mouse)[2] |
|---|
| | Band | 7 D3|7 50.47 cM | Start | 89,779,421 bp[2] |
|---|
| End | 89,862,673 bp[2] |
|---|
|
| RNA expression pattern |
|---|
| Bgee | | Human | Mouse (ortholog) |
|---|
| Top expressed in | - Achilles tendon
- tibial arteries
- corpus callosum
- subcutaneous adipose tissue
- gallbladder
- tibial nerve
- monocyte
- right coronary artery
- C1 segment
- ascending aorta
|
| | Top expressed in | - fetal liver hematopoietic progenitor cell
- interventricular septum
- tibiofemoral joint
- stroma of bone marrow
- left lung lobe
- human fetus
- belly cord
- body of femur
- genital tubercle
- tunica media of zone of aorta
|
| | More reference expression data |
|
|---|
| BioGPS | 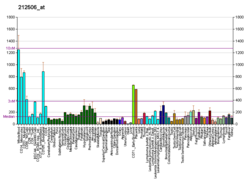
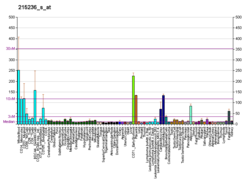
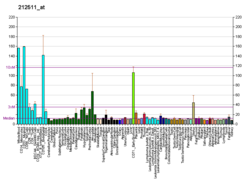 | | More reference expression data |
|
|---|
|
| Gene ontology |
|---|
| Molecular function | - clathrin heavy chain binding
- clathrin adaptor activity
- clathrin binding
- 1-phosphatidylinositol binding
- protein binding
- phospholipid binding
- cadherin binding
- SNARE binding
- amyloid-beta binding
- phosphatidylinositol-4,5-bisphosphate binding
- tau protein binding
- low-density lipoprotein particle receptor binding
| | Cellular component | - clathrin coat of coated pit
- Golgi apparatus
- postsynaptic membrane
- membrane
- neurofibrillary tangle
- neuronal cell body
- clathrin-coated pit
- AP-2 adaptor complex
- presynaptic membrane
- cytoplasmic vesicle
- nucleus
- clathrin-coated vesicle
- intracellular anatomical structure
- vesicle
- perinuclear region of cytoplasm
- cytosol
- intracellular membrane-bounded organelle
- early endosome
- plasma membrane
- cell surface
- intrinsic component of membrane
- clathrin-coated endocytic vesicle
- endosome to plasma membrane transport vesicle
- synaptic vesicle
- extrinsic component of presynaptic endocytic zone membrane
| | Biological process | - regulation of endocytosis
- endocytosis
- iron ion homeostasis
- receptor internalization
- positive regulation of neuron death
- regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process
- negative regulation of gene expression
- regulation of protein localization
- negative regulation of receptor-mediated endocytosis
- positive regulation of transcription, DNA-templated
- endosomal transport
- clathrin coat assembly
- cell population proliferation
- vesicle-mediated transport
- axonogenesis
- hemopoiesis
- clathrin-dependent endocytosis
- negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process
- synaptic vesicle maturation
- positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process
- vesicle cargo loading
- receptor-mediated endocytosis
- dendrite morphogenesis
- positive regulation of amyloid-beta formation
- membrane organization
- iron ion import across plasma membrane
- vesicle budding from membrane
- learning or memory
- positive regulation of GTPase activity
- protein-containing complex assembly
- regulation of vesicle size
- membrane bending
- amyloid-beta clearance by transcytosis
- regulation of amyloid-beta formation
- negative regulation of protein localization to plasma membrane
- clathrin-coated pit assembly
- negative regulation of protein localization to cell surface
- synaptic vesicle budding from presynaptic endocytic zone membrane
- synaptic vesicle endocytosis
| | Sources:Amigo / QuickGO |
|
| Orthologs |
|---|
| Species | Human | Mouse |
|---|
| Entrez | | |
|---|
| Ensembl | | |
|---|
| UniProt | | |
|---|
| RefSeq (mRNA) | |
|---|
NM_001008660
NM_001206946
NM_001206947
NM_007166 |
| NM_001252520
NM_001252521
NM_001252522
NM_001252523
NM_001252524
|
|---|
NM_146194
NM_001360867 |
|
|---|
| RefSeq (protein) | |
|---|
NP_001008660
NP_001193875
NP_001193876
NP_009097
NP_001008660.1 |
| NP_001239449
NP_001239450
NP_001239451
NP_001239452
NP_001239453
|
|---|
NP_666306
NP_001347796 |
|
|---|
| Location (UCSC) | Chr 11: 85.96 – 86.07 Mb | Chr 7: 89.78 – 89.86 Mb |
|---|
| PubMed search | [3] | [4] |
|---|
|
| Wikidata |
| View/Edit Human | View/Edit Mouse |
|

 1hf8: CALM-N N-TERMINAL DOMAIN OF CLATHRIN ASSEMBLY LYMPHOID MYELOID LEUKAEMIA PROTEIN
1hf8: CALM-N N-TERMINAL DOMAIN OF CLATHRIN ASSEMBLY LYMPHOID MYELOID LEUKAEMIA PROTEIN 1hfa: CALM-N N-TERMINAL DOMAIN OF CLATHRIN ASSEMBLY LYMPHOID MYELOID LEUKAEMIA PROTEIN, PI(4,5)P2 COMPLEX
1hfa: CALM-N N-TERMINAL DOMAIN OF CLATHRIN ASSEMBLY LYMPHOID MYELOID LEUKAEMIA PROTEIN, PI(4,5)P2 COMPLEX 1hg2: CALM-N N-TERMINAL DOMAIN OF CLATHRIN ASSEMBLY LYMPHOID MYELOID LEUKAEMIA PROTEIN, INOSITOL(4,5)P2 COMPLEX
1hg2: CALM-N N-TERMINAL DOMAIN OF CLATHRIN ASSEMBLY LYMPHOID MYELOID LEUKAEMIA PROTEIN, INOSITOL(4,5)P2 COMPLEX 1hg5: CALM-N N-TERMINAL DOMAIN OF CLATHRIN ASSEMBLY LYMPHOID MYELOID LEUKAEMIA PROTEIN, INOSITOL(1,2,3,4,5,6)P6 COMPLEX
1hg5: CALM-N N-TERMINAL DOMAIN OF CLATHRIN ASSEMBLY LYMPHOID MYELOID LEUKAEMIA PROTEIN, INOSITOL(1,2,3,4,5,6)P6 COMPLEX
























